MinHash
This chapter is based on Mining of Massive Datasets - The Stanford University InfoLab.
The minHash has a property that the probability for two minimum elements are the same is equal to the Jaccard distance between two arrays.
We want to replace large sets by much smaller representations. We call it as signatures. The important property we need for signatures is that we can compare the signatures of two sets and estimate the Jaccard similaritysignatures alone.
The primary idea of minHash is to have a hash function that is sensitive to distances (Locality Sensitive Hashing - LSH). In other words, if two points are close to each other, the probability that this function hashes them to the same bucket is high. On the contrary, if the points are far apart, the probability that they get hashed to the same bucket is low. There are several distance measures but we opted to use Jaccard distance.
Our sample has 4 sets and 5 elements from the sets as shown in the table below:
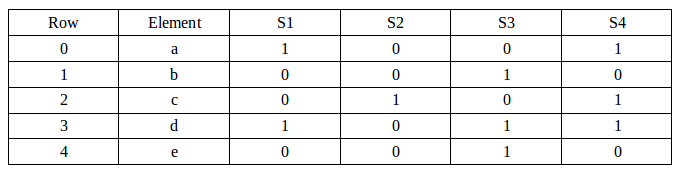
We'll start the following Signature matrix:
The following code simulates the algorithm for computing the signature matrix. In the code, we'll use very simple hash functions which was suggested in the reference I mentioned earlier. Since the steps are well explained in the reference pdf, I'll skip the details.
#!/usr/bin/env python
import sys
def h1(x):
return (x+1)%5
def h2(x):
return (3*x+1)%5
def minhash(data, hashfuncs):
'''
Returns signature matrix
'''
rows, cols, sigrows = len(data), len(data[0]), len(hashfuncs)
# initialize signature matrix with maxint
sigmatrix = []
for i in range(sigrows):
sigmatrix.append([sys.maxint] * cols)
for r in range(rows):
hashvalue = map(lambda x: x(r), hashfuncs)
# if data != 0 and signature > hash value, replace signature with hash value
for c in range(cols):
if data[r][c] == 0:
continue
for i in range(sigrows):
if sigmatrix[i][c] > hashvalue[i]:
sigmatrix[i][c] = hashvalue[i]
return sigmatrix
if __name__ == '__main__':
data = [[1, 0, 0, 1],
[0, 0, 1, 0],
[0, 1, 0, 1],
[1, 0, 1, 1],
[0, 0, 1, 0]]
# print signature matrix
print(minhash(data, [h1, h2]))
Output:
[[1, 3, 0, 1], [0, 2, 0, 0]]
As we can see from the result, our final Signature matrix looks like this:
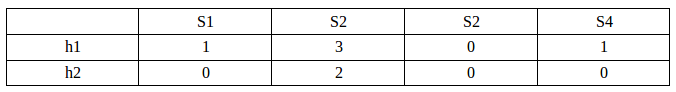
As we've already guessed, it gives us a reasonable output but not exact outcome. For example, the signature matrix thinks Sim(S1,S4)=1 since column 1 and 4 have identical number. However, the true Jaccard similarity is 2/3 from the initial set table. This inaccuracy is due to the fact that we have extremely small size of samples. When we look at (S1,S3), the signature columns differ for h1 but same for h2 which estimates the similarity to be 1/2 while the true similarity is 1/4. For the (S1,S2), the signature matrix estimates the similarity to be 0 which is the correct value.
For LSH with cosine distance, please visit my another post: Locality-Sensitive Hashing (LSH) using Cosine Distance (Cosine Similarity).
Ph.D. / Golden Gate Ave, San Francisco / Seoul National Univ / Carnegie Mellon / UC Berkeley / DevOps / Deep Learning / Visualization